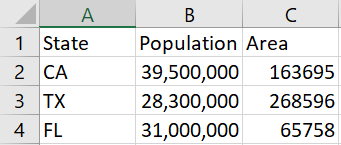
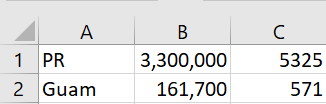
A client emailed me a screenshot of a table rather than pasting the table as text into an email.
I thought about using an LLM to convert it to text, but the table is confidential client information and so I shouldn’t upload it anywhere.
I searched for a command line utility to do OCR and found tesseract. I installed it with
sudo apt install tesseract-ocr libtesseract-dev tesseract-ocr-eng
and ran it with the default settings
tesseract screenshot.png textfile
It worked remarkably well. I had to change a C to a U, but otherwise I didn’t have to add or change any text, but I did have to delete a few extraneous parentheses generated by the software.
I work locally in part out of habit; it was the only way to work when I started using a computer. It has numerous advantages, such as being able to keep working when a hurricane knocks out my internet connection, but above all it is private.
I pay more attention to privacy than is convenient because I work in data privacy. And aside from my privacy, I have to protect our clients’ privacy.
Update: According to the comments, ChatGPT uses tesseract. Assuming that’s true, using tesseract directly is better than ChatGPT because it does exactly what you want. No ambiguity as far as what expected. No potential for tinkering with your results before you see them.